To ensure they give birth to a healthy baby, pregnant mothers will take the necessary precautions, such as eating a balanced diet or avoiding exposure to cigarette smoke. This is because there appears to be a link between exposure to environmental factors early in development and disease in adulthood. One explanation for this relationship may involve epigenetic modifications, such as DNA methylation of transposable elements.
The study featured in the article, Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development, sought to explore the effects of chemical exposure, specifically bisphenol A, during early development on the epigenome and whether or not these effects could be counteracted by nutrient supplementation.
Epigenetic modifications established during early development create epialleles, or alleles that would be identical if not for methylation differences. One example of an epiallele is the Agouti gene in the yellow agouti mouse. The Avy allele of this gene resulted from the insertion of a retrotransposon into the 5' end of the gene. Methylation of cytosines in cytosine-guanine (CpG) dinucleotide sites in and near the Avy gene results in a wide variation in coat color, ranging from yellow (unmethylated) to pseudoagouti (methylated).
Bisphenol A (BPA) is a chemical used in the manufacture of polycarbonate plastics. It can be found in products such as food and beverage containers and baby bottles. Although reviews of the BPA literature have conflicting conclusions as to its potential as a human health risk, rodent studies have found that exposure to BPA, either as an embryo or fetus or as a newborn, is associated with higher body weight, increased breast and prostate cancer, and altered reproductive functions. The current study looked at the effect on the epigenome of offspring of maternal BPA exposure alone and in combination with nutritional supplements.
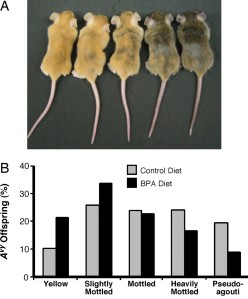 The study showed that maternal BPA exposure shifted the coat color distribution of genetically identical heterozygous offspring towards the yellow coat color phenotype. Twenty-one percent of offspring exposed to BPA during development were yellow compared with ten percent of offspring with no BPA exposure. Bisulfite treatment and sequencing was used to measure the DNA methylation at nine CpG sites in the promoter region of the Avy allele. The offspring that were exposed to BPA had 27+/- 2.8% methylation across the nine sites compared with 39 +/- 2.6% in the offspring without BPA exposure. The sites that exhibited the greatest difference in methylation appear to be important in modifying chromatin structure and Agouti gene expression. Early stem cell development must to be sensitive to BPA exposure since the patterning of DNA methylation at the Avy locus was similar in tissues from three of the germ layers (ectoderm, mesoderm, and endoderm).
The study showed that maternal BPA exposure shifted the coat color distribution of genetically identical heterozygous offspring towards the yellow coat color phenotype. Twenty-one percent of offspring exposed to BPA during development were yellow compared with ten percent of offspring with no BPA exposure. Bisulfite treatment and sequencing was used to measure the DNA methylation at nine CpG sites in the promoter region of the Avy allele. The offspring that were exposed to BPA had 27+/- 2.8% methylation across the nine sites compared with 39 +/- 2.6% in the offspring without BPA exposure. The sites that exhibited the greatest difference in methylation appear to be important in modifying chromatin structure and Agouti gene expression. Early stem cell development must to be sensitive to BPA exposure since the patterning of DNA methylation at the Avy locus was similar in tissues from three of the germ layers (ectoderm, mesoderm, and endoderm).
Additionally, results from a statistical approach, known as mediational regression analysis, suggest that BPA exposure does not directly regulate coat color. Instead, coat color is determined by the methylation at the Avy gene. BPA exposure just so happens to affect methylation and therefore indirectly affects coat color.
The study investigated the relationship between BPA exposure and maternal nutritional supplements that are methyl donors, such as folic acid. Previous studies suggest that these supplements will counteract the hypomethylation effect of BPA exposure. Female mice that received a BPA diet supplemented with methyl donors or genistein (not a methyl-donating compound) produced offspring that exhibited the coat color distribution found in offspring from females that were not exposed to BPA. These findings suggest that maternal nutritional supplementation, with either methyl-donors or non-methyl-donors, counteracts the hypomethylation caused by maternal BPA exposure.
BPA exposure is linked to modifications of the epigenome and disease pathologies that are passed down through the germ line. This study’s findings suggest that nutrition interventions during early development have the potential to reduce disease susceptibility in adulthood. Be sure to thank your mom for eating healthy during her pregnancy!



