Saturday, June 9, 2012
Type II Diabetes: It’s Not Just a Human Problem!
Tuesday, May 1, 2012
Are You What Your Mom Ate?
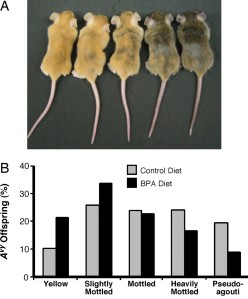 The study showed that maternal BPA exposure shifted the coat color distribution of genetically identical heterozygous offspring towards the yellow coat color phenotype. Twenty-one percent of offspring exposed to BPA during development were yellow compared with ten percent of offspring with no BPA exposure. Bisulfite treatment and sequencing was used to measure the DNA methylation at nine CpG sites in the promoter region of the Avy allele. The offspring that were exposed to BPA had 27+/- 2.8% methylation across the nine sites compared with 39 +/- 2.6% in the offspring without BPA exposure. The sites that exhibited the greatest difference in methylation appear to be important in modifying chromatin structure and Agouti gene expression. Early stem cell development must to be sensitive to BPA exposure since the patterning of DNA methylation at the Avy locus was similar in tissues from three of the germ layers (ectoderm, mesoderm, and endoderm).
The study showed that maternal BPA exposure shifted the coat color distribution of genetically identical heterozygous offspring towards the yellow coat color phenotype. Twenty-one percent of offspring exposed to BPA during development were yellow compared with ten percent of offspring with no BPA exposure. Bisulfite treatment and sequencing was used to measure the DNA methylation at nine CpG sites in the promoter region of the Avy allele. The offspring that were exposed to BPA had 27+/- 2.8% methylation across the nine sites compared with 39 +/- 2.6% in the offspring without BPA exposure. The sites that exhibited the greatest difference in methylation appear to be important in modifying chromatin structure and Agouti gene expression. Early stem cell development must to be sensitive to BPA exposure since the patterning of DNA methylation at the Avy locus was similar in tissues from three of the germ layers (ectoderm, mesoderm, and endoderm).Not So Identical
Friday, June 10, 2011
The benefits of RNA in curing cancer
Neils Pierce of the California Institute of Technology (Caltech) has developed a new method of killing cancer cells. Pierce’s main goal was to develop a therapy that did not result in side effects and create an innovative way of killing cancer cells using small RNA molecules. Pierce and his colleagues used a method using two types of small conditional RNA molecules, which are 30 base pairs in length. According to the researchers, one of the RNA molecules “is designed to be complementary to, and thus to bind to, an RNA sequence unique to a particular cancer cell.” The RNA hairpin then changes form by opening and exposing a sequence that can spontaneously bind to the second type of RNA hairpin that will bind to the cancer mutation; this process continues from one hairpin to the next.
“In this way, detection of the RNA cancer marker triggers the self-assembly of a long double-stranded RNA polymer.” In order to search for long double-stranded viral RNA in the human cells, they used a protein known as protein kinase R (PKR). If the protein is able to find the long double-stranded RNA in the cell, PKR will trigger the cells to undergo apoptosis, a cell death pathway to destroy the cell. The results of their study showed that small RNAs were able to “trick” the cancer cells to self-destruct.
Pierce concluded that small RNA molecules were advantageous due to the fact that they were useful in the diagnosis and treatment of the disease “one cell at a time.” However, further studies will need to be conducted in order to see if the small RNAs can be useful for the diagnosis in human patients.” Pierce’s approach to finding a way to “program” cancer cell death is beneficial to those diagnosed with cancer due to the fact that these patients would not have to be overwhelmed with the additional side effects that come about from cancer drugs and therapy.
Citation: California Institute of Technology. "Scientists create new process to 'program' cancer cell death." ScienceDaily, 8 Sep. 2010. Web. 28 May 2011.
Thursday, June 9, 2011
Epigenetic and Breast Cancer Sub-types
Tuesday, June 7, 2011
New Target for Breast Cancer?
Further analysis revealed that the protein encoded by C6ORF211 was expressed mainly in the cytoplasm. C6ORF211 was shown to drive the growth of tumor. In a proteomic screen the protein has been found to interact with SAP18, a Sin3A-associated cell growth inhibiting protein. This reported interaction was hypothesized as one of the reasons that there was a suppression of proliferation in cultured cells where C6ORF211 was knocked down. Contrarily, a high level of activity of C6ORF97 predicted an improvement of “disease-free survival” in tamoxifen-treated dataset, independently from ESR1. The gene was a good predictor of response to tamoxifen. Less was known about C6ORF96, but it was being researched by the team.
Tamoxifen works by competitively blocking binding of oestrogen to receptors and therefore decreasing the transcriptional activation level of genes required for tumor growth. Tamoxifen is not shown to efficiently affect the activity of the new discovered genes, thus opening up a possible synergistic drug treatment for breast cancer along with the current treatment. Professor Mitch Dowsett, who lead the team at the Breakthrough Breast Cancer Research Centre at the ICR, added:
"This research is exciting because it shows that while the oestrogen receptor is the main driver of hormonal breast cancer, there are others next door to it that also appears to influence breast cancer behavior. We now need to better understand how they work together and how we can utilize them to save lives of women with breast cancer."
Hopefully the discovery of the new genes will help us to develop new drugs can cure breast cancer. The problem with the current treatment is that he tumors develop resistance overtime and may come back after the surgery and spread to another place. Perhaps with better and more advanced technologies, cancer can be treated as a curable disease.
Citation:
Scientist discover three genes link to breast cancer. Thursday, 5th May 2011. Mackenzie, Carla.
<http://www.figo.org/news/
C6ORF211 Genes Catalyze the Growth of Tumor in Breast. Wednesday, 4th May 2011. Wilkins, Dave.
<http://topnews.us/
Three gene discovery may lead to new breast cancer treatments. 4th May 2011. Kraft, Sy.
<http://www.medicalnewstoday.
Oestrogen receptors and breast cancer. Elledge, Richard M. Osborne, C Kent. University of Texas Health Science Center, San Antonio, TX 78284-7884, USA.
<http://www.bmj.com/content/
ESR1 is co-expressed with closely adjacent uncharacterized genes spanning a breast cancer susceptibility locus at 6q25.1. Anita K. Dunbier, Helen Anderson, Zara Ghazoui,Elena Lopez-Knowles, Sunil Pancholi, Ricardo Ribas,Suzanne Drury, Kally Sidhu, Alexandra Leary, Lesley-Ann Martin, Mitch Dowsett. May 12th, 2011. London, United Kingdom.
<http://www.plosgenetics.org/
Friday, June 3, 2011
Autism, Where Did It Come From?
Autism is a developmental disorder that impairs social behavior and communication in children before the age of 3. While the current estimate of a sibling recurrence risk is at 15%, the population of children with an autism spectrum disorder is about 1 per 150. How has autism become so common within these young children? Geneticists have found a couple of important genes called NHE9 and DIA1 as possible disruptions to cause the disorder.
A study done by Morrow et. al, investigated the coding regions of NHE9, a nearby gene encoding a membrane protein that exchanges intracellular H-ions for sodium. It was sequenced to find a loss of function mutation in a nonconsanguineous family. They also found a gene called DIA1 (deleted in autism1), an uncharacterized protein, to be completely removed from chromosome 3q.
Morrow et. al, used DNA microarrays to study various consanguineous families from the Middle East and were able to identify common inherited regions of affected individuals with homozygous segments. These individuals were completely deficient for the genes in the deleted intervals and these regions were predicted to cause the autism spectrum disorder in the family.
These scientists also found similar loss of function mutations causing an epileptic phenotype in mice. These mutations also caused a phenotype with autistic symptoms and epilepsy in the related NHE6 gene. Mice can be tested for autism by monitoring the reduction of social response to other individuals. They can also be startled by an auditory signal in order to measure its response, frozen time, and its force applied to the floor. Combining these findings support dysregulation of NHE9 as a contributing or casual factor in a family with affected individuals.
Overall, this study points out some important clues towards autism susceptibility. Since autism is a neurodevelopmental disorder, the genes mentioned above have expression changes due to stimulation of neuronal activity. As the brain develops after birth, synapses mature as a function of experience-dependent neuronal activity and of the gene-expression that follows. Scientists have been able to establish dysregulation of synaptic development as an idea for autism research. Although more studies are necessary to confirm this idea, the possibility that dysregulation of these genes results in synaptic development disruption is a fascinating hypothesis.
Sutcliffe, James S. Insights into the Pathogenesis of Autism. Science 11 July 2008: 208- 209. [DOI:10.1126/science.1160555]
Anders J, Baxter B, Pi C, Dunn C, Fahimi F, Yamdagni N. Novel Measures of Mouse Social Behavior. December 2004. http://homepages.cae.wisc.edu/~bme402/mouse_stress/reports/Final_Paper.pdf






